Characterization of Inner Cavity Lining of Proteins
Files You Get
- Original PDB
- Another PDB with "-inner_residues.pdb" added after the original filename
- A graph with the Diameter-Profile of the Inner Surface of the submitted structure
- A graph with the Volume-Profile of the Inner Surface of the submitted structure
- A file listing all the inner residues in the following format "<Residue Number>.<Chain ID>"
- A Log file indicating if any errors were encountered, and what can be done to resolve it
- In case the option to calculate the conservation scores was marked as YES, the following files are also provided
- A PDB with the conservation scores written in the b-factor (Temp. factor) column
- A value of "10" means that the corresponding atom does not lie on the detected inner surface
- For all the other atoms, the b-factor column contains the corresponding conservation score for its residue
- Text files containing the conservation scores of all the residues, a different file for every distinct chain in the submitted PDB
- The Multiple Sequence Alignment (MSA) input and output files
- The input (The file containing the querry sequences) and output files containing sequence homologues as detected by Psi-Blast using the "nr" database
How to Visualize the Inner Surface
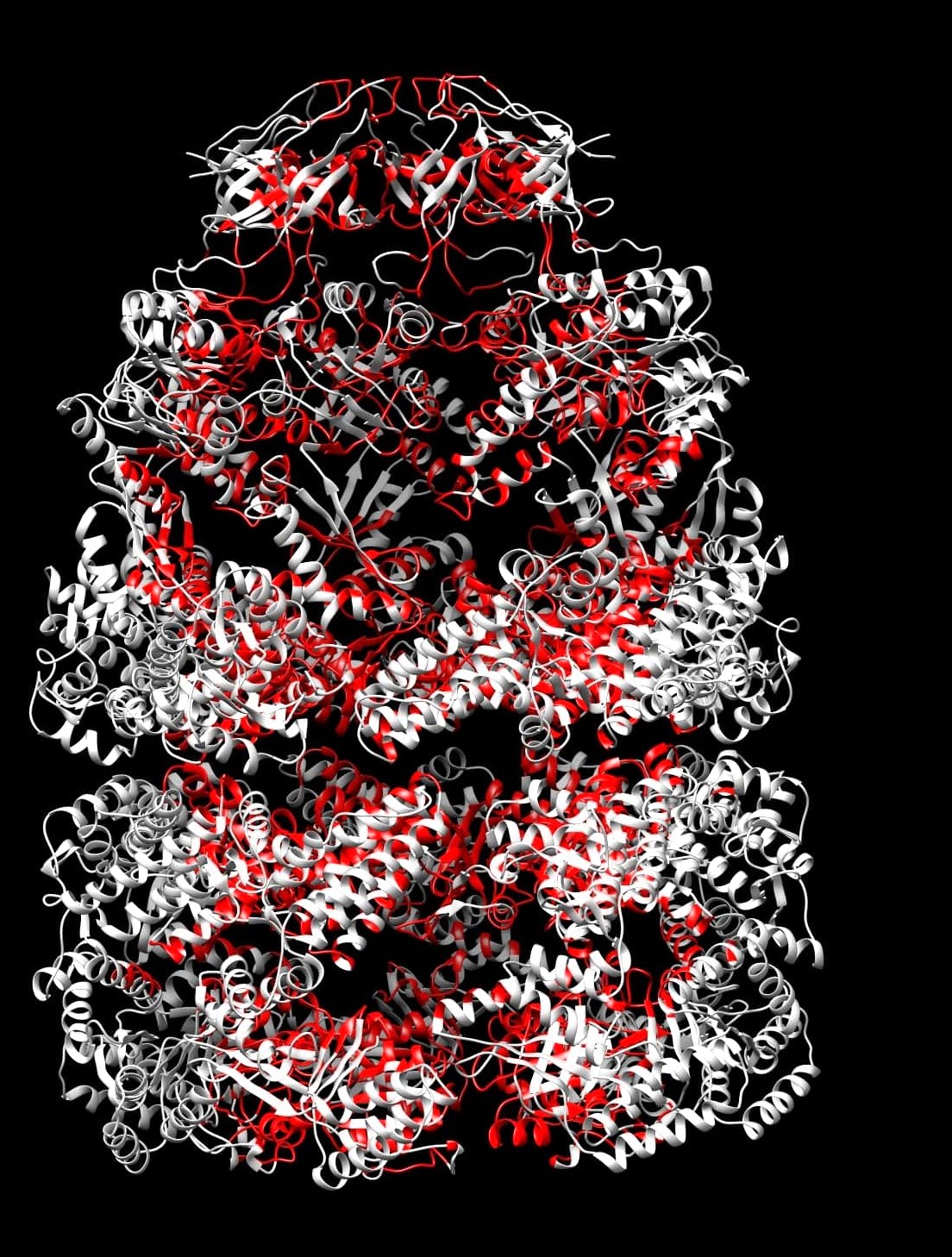
-- Open the PDB with "-inner_residues.pdb" added after the original filename in any molecular visualizer of your choice
-- The information for an atom being on the inner surface is stored as the b-factor values of the atoms in this PDB. An atom with a b-factor of 9999 is determined to be on the inner surface of the structure while those with a value of 0 are not
-- For viewing the inner surface in an atomistic fashion, render the PDB as atoms and color them according to their b-factor
(It's as simple as that)
-- For viewing the inner surface as ribbons (Just like the image to your left) or in any other format, render the image in the required format and color all the residues with a single color. Open the file having "-residue_list.dat" after the PDB name. This file contains all the residues on the inner surface in the format "Residue_Number.Chain_ID (residue number dor chain ID)" (Beware of the extra "," at the end of the file). Select all the residues in this file and color them with a color other than the one used a little while ago in this section. And Voilla, There you have it!!